An observed toxicological effect begins with a molecular initiating event (MIE). Examples of toxicological effects can be liver tumor formation, cell death, abnormal growth, or cell malformation. Toxicogenomic studies help us determine these MIEs by identifying the [suspected] subsets of genes that would lead to their trigger. These suggested subsets of genes that might be responsible for bioactivity can be confirmed or infirmed after additional in vitro studies. While a compound might trigger an initiating event, it doesn’t necessarily need to cascade to an adverse outcome as the body might trigger other actions to prevent a negative effect.
There are many software tools available to analyze data from toxicogenomic studies. ScitoVation is a pioneer in developing these software tools including BMD. While the use of these software tools can be relatively simple to use, the complexity from these studies lies in the interpretation of the data. The value we bring to clients is the multi-disciplinary expertise of our team of bioinformaticians, biologists and toxicologists. Additionally, our clients find that our expertise with a broad range of chemicals and the number of toxicogenomic studies we conducted or analyzed bring depth to our interpretation of these data and their acceptance by other stakeholders.
Transcriptomic Case Studies
Use of Transcriptomics as a read-across
“Read across” is the process of filling toxicology data gaps using information about compounds with similar structure. It is common to use structure-based approaches in risk assessments and registration applications. The principal of read across is that compounds with similar structural features will have similar biological activity, and therefore will elicit similar toxicity. A biological read across, by contrast, directly measures biological activity driven by exposure. The rational is that compounds with similar bioactivity profiles are more likely to lead to the same toxicity effects (Figure 1A). In toxicogenomics, the biological activity that we compare in a read-across is the genetic response to a similar compound.
For a REACH registration, a client had a mixture of isomers (chemicals with the same formula that differs only by their shape) and wanted to determine if the individual compounds were similar in their toxicological response to the mixture. We performed a toxicogenomic study that compared the genetic response of each of the isomers and the mixture. From our study, we did not find any of the isomers to be different than the mixture.
We believe that a potential case for toxicogenomic tools is to augment a read-across argument for REACH registration. For illustration, consider a hypothetical class of chemicals that differ by substitution of an alkyl group (Figure 1). Many compounds in the class have been well studied, and there is a general understanding that toxicity in a known target organ increases with chain length. A client has a target compound of interest for which there is no toxicity data (Target Compound). The target compound has an alkyl group one carbon longer than a short chain length analog with no toxicity (Source Compound A- Figure 2).
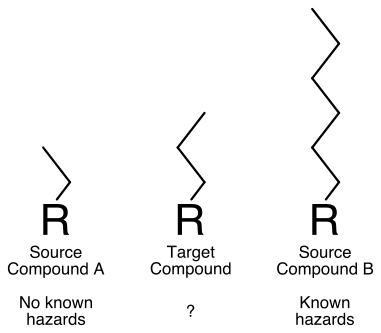
Figure 1. Hypothetical Read Across
To strengthen the structure-based read across that would be the basis for the registration, the study involves selection of a suitable cell line related to the target tissue toxicity. Here, we would opt in this scenario to use two data-rich source compounds—one short chain (Source Compound A) and one long chain (Source Compound B)—in addition to the target compound. Cells would be exposed to each of the three compounds in concentration-response format. RNA would be collected and sequenced. The resulting data would be processed using our established bioinformatic pipeline, which would score the pairwise similarity between the three compounds. If the target bioactivity is similar to the short-chain source compound, this provides additional evidence for its use as a read across anchor.
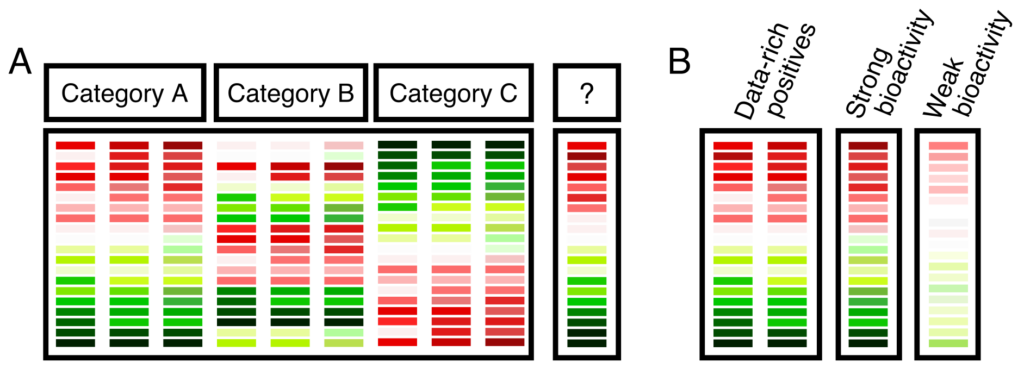
Figure 2. Machine Learning MOA TGX Composite
Using Transcriptomic Points of Departure to Determine Compound Toxicity
Conventional concepts of points of departure for adverse effects (e.g., NOAEL/LOAEL) do not translate directly to gene expression data. The benchmark dose approach offers a robust alternative method, and has been adapted to transcriptomics. Benchmark Dose is a data driven mathematical approach to determine a dose at which a significant change in response is detected. It is applicable to continuous data such as gene expression data where changes in the abundance of gene transcripts can be detected for all genes expressed in a cell using fluorescence detection (as in microarray technology) or via counts of sequenced transcripts per gene (as with RNA-Seq or BioSpyder’s TempO-Seq ligation system).
Chemicals with different potencies lead to apical effects at different exposure levels (i.e., have different points of departure, POD). It has been similarly established that compounds with lower points of departure also induce transcriptomic changes at lower concentrations. This observation has led to the concept of the transcriptomic point of departure as a proxy for compound toxicity. The impact of a compound on the transcriptome can be quantitatively determined and used to compare potency (Figure). We are using this concept with clients in applications related to compound screening and down-selection. There are also potential applications in product stewardship, such as alternatives assessment.
Increasing Predictability with BioMarkers Time and Cost Savings
A potential of transcriptomic data is to derive a single or small number of highly sensitive genes or transcriptional markers (e.g. single nucleotide changes or SNPs within a gene) that have high specificity for a particular adverse outcome. An example of this is a relatively small number of genes, isolated by predictive modeling of whole transcriptomic changes, which in aggregate produce changes in gene expression that are highly predictive of a genotoxic response to exposure. This concept has been applied to a gene panel where human TK6 cells are exposed to a chemical or environmental perturbagen, and expression of 65 genes is rapidly and inexpensively measured by qPCR, and mathematical models of the resolved expression patterns indicates a chemical probability of inducing genotoxicity.
Using Biomarkers to Identify Mode of Action (MoA)
A client had a compound that was observed to cause cancer in animal studies. The client wanted to know if the findings from these animal studies were relevant to humans. We conducted studies where we compared the genetic response from the rats and a relevant human cell line. The data from the rat showed a pattern known as Ppar alpha. For this pattern, it is known to cause cancer in rats but not in humans.
Transcriptomic Resources:
We conducted several papers involving toxicogenomic data and actively collaborate with other researchers on this topic.
Our researchers developed a curated list of trascriptomics papers as a start for those who are interested in the topic.
MoAViz: Our own software tool for analyzing and visualizing data from toxicogenomic studies
BMD: This software is currently maintained by NIEHS. It was developed at the Hamner, the predecessor organization of ScitoVation.
Key manuscripts referenced by Mel Andersen in his October 8, 2020 webinar (View the webinar on-demand here)
Chemical-specific case studies
Formaldehyde
- Thomas, R. S. et al. (2007) A method to integrate benchmark dose estimates with genomic data to assess the functional effects of chemical exposure. Toxicol Sci 98, 240-248, DOI: 10.1093/toxsci/kfp233
- Andersen, M. E., Clewell, H. J., 3rd, Bermudez, E., Willson, G. A. & Thomas, R. S. (2008) Genomic signatures and dose-dependent transitions in nasal epithelial responses to inhaled formaldehyde in the rat. Toxicol Sci 105, 368-383, DOI: 10.1093/toxsci/kfn097
- Andersen, M. E. et al. (2010) Formaldehyde: integrating dosimetry, cytotoxicity, and genomics to understand dose-dependent transitions for an endogenous compound. Toxicol Sci 118, 716-731, DOI: 10.1093/toxsci/kfq303
Chloroprene
- Thomas, R. S. et al. (2013) Cross-Species Transcriptomic Analysis of Mouse and Rat Lung Exposed to Chloroprene. Toxicological Sciences 131, 629-640, DOI: 10.1093/toxsci/kfs314
Vanadium Pentoxide
- Black, M. B., et al. Using gene expression profiling to evaluate cellular responses in mouse lungs exposed to V2O5 and a group of other mouse lung tumorigens and non-tumorigens. Regul. Toxicol. Pharmacol. 73: 339 (2015), DOI: 10.1016/j.yrtph.2015.07.017
- Thomas, R. S. et al. (2009) Use of short-term transcriptional profiles to assess the long-term cancer-related safety of environmental and industrial chemicals. Toxicol Sci 112, 311-321, https://www.ncbi.nlm.nih.gov/pubmed/19776212, DOI: 10.1093/toxsci/kfp233
Methylene chloride
- Andersen, M. E., et al. Combining transcriptomics and PBPK modeling indicates a primary role of hypoxia and altered circadian signaling in dichloromethane carcinogenicity in mouse lung and liver. Toxicol. Appl. Pharmacol. 2017, 332, 149-158, DOI: 10.1016/j.taap.2017.04.002
- See also public comments in EPA-HQ-OPPT-2019-0437 by Mel Andersen
Styrene
- Andersen, M. E., et al. Assessing molecular initiating events (MIEs), key events (KEs) and modes-of-action (MOAs) for styrene responses in mouse lungs using whole genome gene expression profiling following 1-day and multi-week exposures. Toxicol. Appl. Pharmacol. 2017, 335, 28-40, DOI: 10.1016/j.taap.2017.09.015
- Andersen, M. E., et al. Strain-related Differences in Mouse Lung Gene Expression Over a Two-year Period of Inhalation Exposure to Styrene: Relevance to Human Risk Assessment. Regulatory Toxicology and Pharmacology. 2018, 96: 153-166, DOI: 10.1016/j.yrtph.2018.05.011
Acetamide
- Nault, R. et al. A toxicogenomic approach for the risk assessment of the food contaminant acetamide. Toxicol. Appl. Pharmacol. 2020, 388: 114872, DOI: 10.1016/j.taap.2019.114872
Perspectives on the use of transcriptomics to inform mode of action
- Andersen, M. E., et al. Application of Transcriptomic Data, Visualization Tools and Bioinformatics Resources for Informing Mode of Action. Current Opinion in Toxicology. 2018, 9: 21-27, DOI: 10.1016/j.cotox.2018.05.003
