Key Takeaways:
Benchmark dose (BMD) analysis offers a cost efficient and rapid means of categorizing the risk associated with a chemical. Using BMD data, margins of exposure can be computed to compare exposure scenarios against exposures that cause transcriptomics changes. A practical application for BMD is as a screening tool to select from a group of chemicals based on their potential hazard. Additionally, ScitoVation has developed tools to explore possible modes of actions (MoAs) based on the subsets of genes that are expressed at different dosage. The information on possible MoA for a chemical can help inform the direction and design of follow-up studies for a selected chemical. These methods are also currently under adoption by the U.S. EPA in their HTTr program (High Throughput Transcriptomics Concentration-Response Screening) using high throughput transcriptomic count data (BioSPyder TempO-Seq) from in vitro whole transcriptomic arrays.[*]
“The dose makes the poison” is a fundamental creed of Toxicology, capturing the importance of dose-response modeling in understanding a chemical’s risk to human health. As such, a major endeavor among toxicology practitioners is evaluating what levels of a chemical should be considered safe, versus what levels have potential to cause adverse health effects. The dose separating these domains is called the point of departure (POD). Traditionally, human health risk assessment has defined and extrapolated a POD from the “no observed adverse effect level” (NOAEL) in animal toxicology studies1. However, this approach has several limitations, including high dependence on dose selection and sample size, as well as the inability to appropriately account for study quality or uncertainty and make cross-study comparisons.The Benchmark Dose (BMD) estimation approach, a computational method, overcomes these limitations because it is based on mathematical model fitting to observed dose-response in vivo or in vitro exposure data from genomic studies. 2,3
Beyond the normal baseline response in controls, the BMD is the chemical exposure or dose that corresponds to a predefined change in the response rate of an adverse event, or the Benchmark Response (BMR).
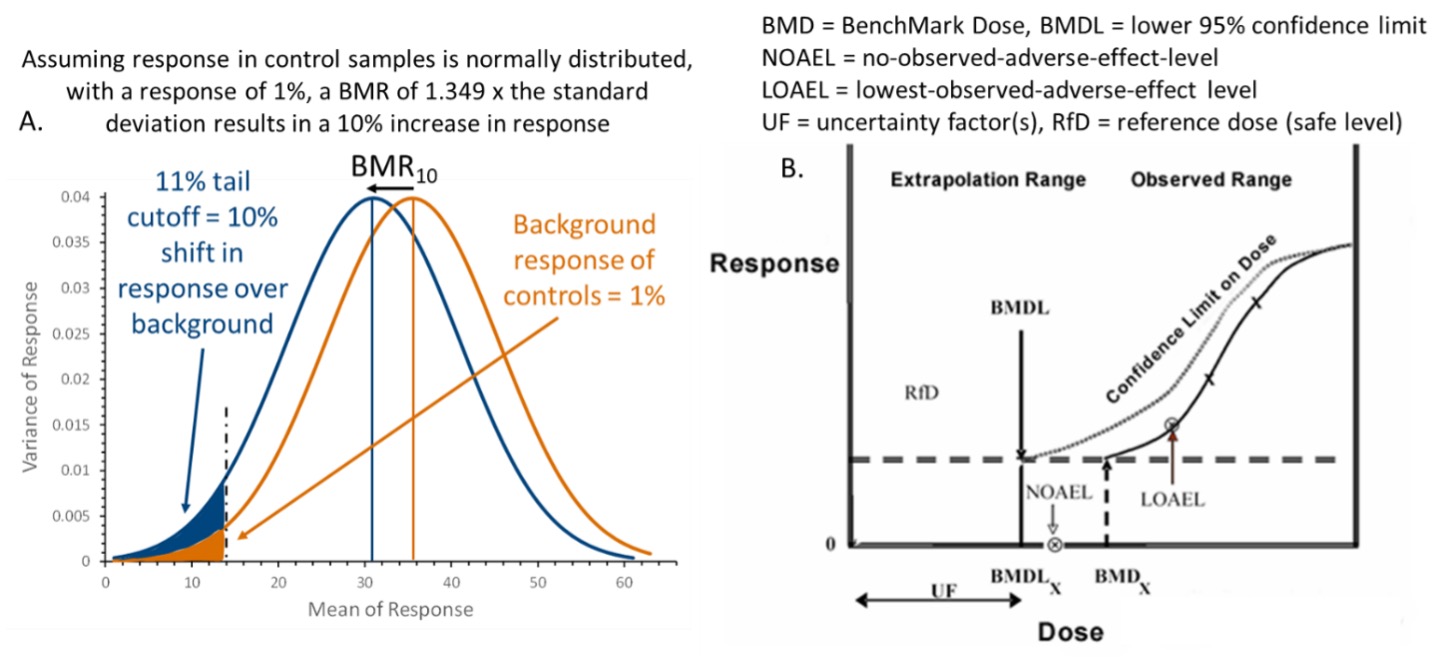
Once a BMR has been selected, a dose-response curve is fit, and the BMD computed.4 The lower confidence limit, or BDML, is usually selected as the POD for risk assessments as it explicitly accounts for the variability in the experiment to produce a conservative estimate. This approach is advantageous in that it can potentially consider any dosing level, studies the dose-response curve in totality, and applies a consistent response level across studies and study types. An acceptable safe dose (e.g., a Reference Dose) is determined from this POD with the application of uncertainty factors such as interspecies conversion uncertainty for data derived from in vivo animal testing, uncertainty due to limitations of available exposure data, or other sources of variability.
Extending BMD to whole genome transcriptomic data
In 2007, while at the Hamner Institutes (predecessor to ScitoVation), Dr. R. Thomas and colleagues extended the concept of BMD to groundbreaking analyses of gene expression data.5 BMD computation was applied to whole transcriptome gene expression response (microarray or RNA-Seq data using whole cell mRNA) by fitting a variety of dose-response curves to every probe in a microarray dose-response experiment or every gene in an RNA-Seq experiment. The aim was to estimate the POD at which transcriptomic change happens. The rationale behind this approach is that changes in gene expression must precede the observed apical changes in phenotype (e.g., cancer, liver hypertrophy) and thus dose response changes in gene expression may be a protective surrogate for an apical endpoint without the need for long term in life studies. Studies using in vivo transcriptomics for several NTP 2-year cancer endpoint compounds demonstrated a high degree of correlation of transcriptomic based BMDs with their NTP apical end point BMD.6 BMDL values, from transcriptomic data or other dose response data can then be adjusted by appropriate uncertainty factors to derive an RfD that is protective of human health. Shown below is an example for nitroguanidine which is used as a propellent in automotive air bags and as a fertilizer and which shows fetal and developmental toxicity in rats.7 The BMD based estimates are shown relative to the combinatorial IRIS determined RfD based on multiple data studies.
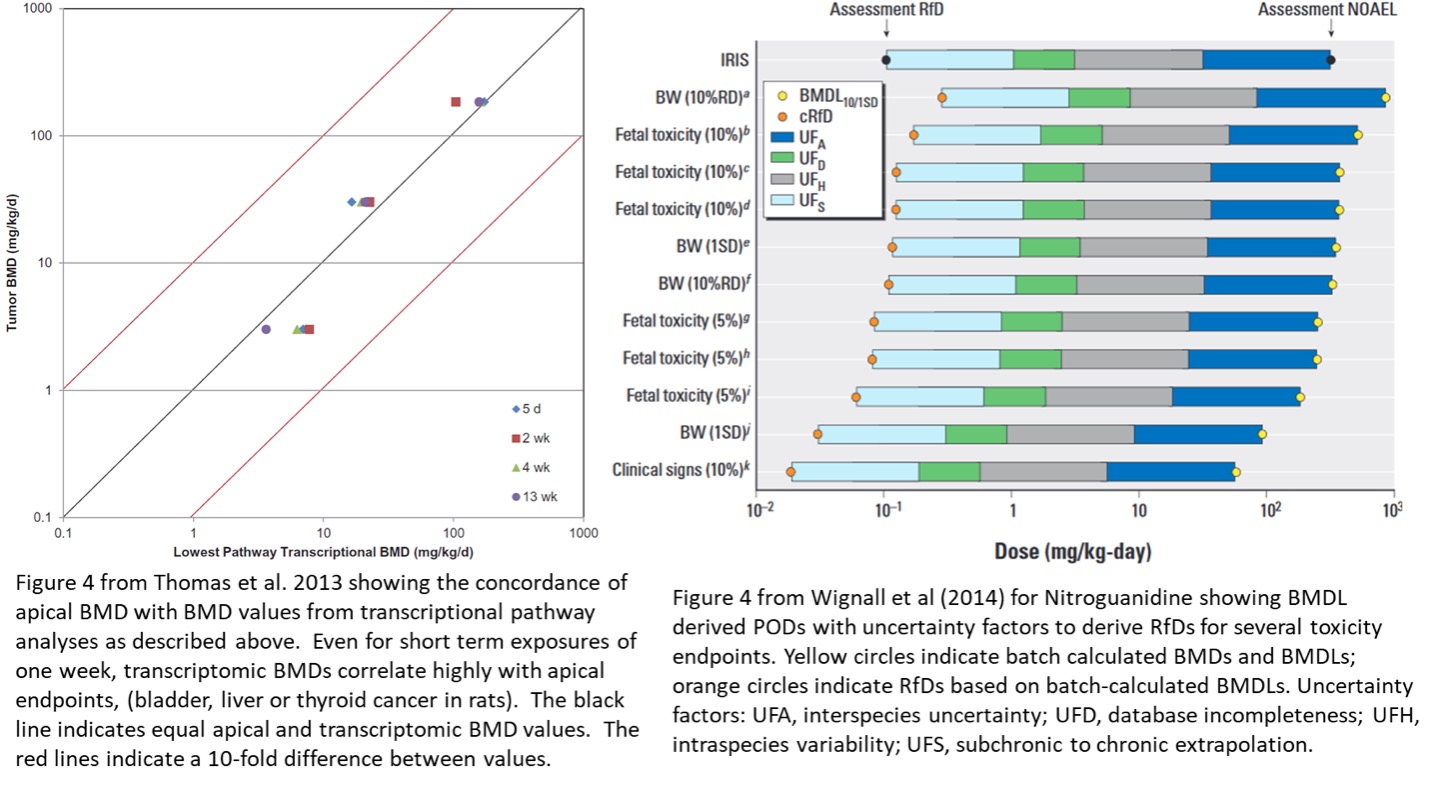
This transcriptomic BMD approach was released in the BMDExpress software now under the stewardship of the US NIEHS National Toxicology Program (NTP) as BMDExpress2.[†] The BMD results could then be applied to cellular functional ontology (i.e., databases of biochemical process or pathway) data. This then led to the application of transcriptomic-based risk analysis of genetic changes from chemical exposure and their associated functional ontology categorization.6,8 Cellular functional pathway BMD-based PODs correlate well (r>0.9) with their conventional associated apical endpoint PODs, even for transcriptomic dose-response exposures of only one week.6 That is the observed POD from transcriptomics studies correlate well with the endpoints observed in animal studies (e.g., observed tumors). Ultimately, extending BMD to whole genome transcriptomic data allows us to design highly valid studies focusing on cellular pathways that save our customers time and money.
Why Conduct Benchmark Dose Analysis Early in the Safety Assessment Process?
Benchmark dose analysis offers a cost efficient and rapid means of categorizing the risk associated with a chemical. Using BMD data, margins of exposure can be computed to compare exposure scenarios against exposures that cause transcriptomics changes. A practical application for BMD is as a screening tool to select from a group of chemicals based on their potential hazard. Additionally, ScitoVation has developed tools to explore possible modes of actions (MoAs) based on the subsets of genes that are expressed at different dosage. The information on possible MoA for a chemical can help inform the direction and design of follow-up studies for a selected chemical. These methods are also currently under adoption by the U.S. EPA in their HTTr program (High Throughput Transcriptomics Concentration-Response Screening) using high throughput transcriptomic count data (BioSPyder TempO-Seq) from in vitro whole transcriptomic arrays.[‡]
References
- Dorato, M. A. & Engelhardt, J. A. The no-observed-adverse-effect-level in drug safety evaluations: use, issues, and definition(s). Regul Toxicol Pharmacol 42, 265-274, doi:10.1016/j.yrtph.2005.05.004 (2005).
- Haber, L. T. et al. Benchmark dose (BMD) modeling: current practice, issues, and challenges. Crit Rev Toxicol 48, 387-415, doi:10.1080/10408444.2018.1430121 (2018).
- Adeleye, Y. et al. Implementing Toxicity Testing in the 21st Century (TT21C): Making safety decisions using toxicity pathways, and progress in a prototype risk assessment. Toxicology 332, 102-111, doi:10.1016/j.tox.2014.02.007 (2015).
- Davis, J. A., Gift, J. S. & Zhao, Q. J. Introduction to benchmark dose methods and U.S. EPA’s benchmark dose software (BMDS) version 2.1.1. Toxicol Appl Pharmacol 254, 181-191, doi:10.1016/j.taap.2010.10.016 (2011).
- Thomas, R. S. et al. A method to integrate benchmark dose estimates with genomic data to assess the functional effects of chemical exposure. Toxicological Sciences 98, 240 (2007).
- Thomas, R. S. et al. Temporal concordance between apical and transcriptional points of departure for chemical risk assessment. Toxicol Sci 134, 180-194, doi:10.1093/toxsci/kft094 (2013).
- Wignall, J. A. et al. Standardizing benchmark dose calculations to improve science-based decisions in human health assessments. Environ Health Perspect 122, 499-505, doi:10.1289/ehp.1307539 (2014).
- Thomas, R. S. et al. Application of transcriptional benchmark dose values in quantitative cancer and noncancer risk assessment. Toxicological Sciences 120, 194 (2011).
