Key Takeaways
Transcriptomics coupled with in vitro experimental design can provide a cost-efficient way for generating hypotheses on possible mode of actions that can be confirmed in follow-up studies. Additionally, the information on the doses at which transcriptomics change happen from these studies can be used to classify risk level and help guide design of future studies. In sum, transcriptomics studies are a useful tool for quantifying the risk of a compound early in the process. At the same time, the information from transcriptomics studies can lead to both time and cost savings in follow-up studies.
Transcriptomics as a means of establishing compound mode of action
Toxicologists use a variety of experimental systems to explore how chemicals might affect people. The means by which a chemical causes toxicity in a biological system—its mode of action—can have profound implications for its safety in humans. For example, compounds that interact directly with DNA have potential to cause mutations at low concentrations that could lead to cancer. These “genotoxic” compounds are subject to different rules than other compounds. Similarly, some chemical modes of action are known to impact children more than adults. Compounds with these modes of action are also treated with more scrutiny.
ScitoVation scientists have a long history of helping our clients better understand the modes of action of their compounds of interest. A particularly valuable approach that has increased stakeholder confidence in these studies has been transcriptomics, which allows us to simultaneously measure how every gene in an experimental system responds to an experimental treatment. Because biological systems actively adapt to their environments, the nature of this transcriptomic response is highly indicative of the compound’s mode of action. We have used this concept to test hypotheses about how compounds work.
A case example for transcriptomics-based mode of action analysis: human relevance of rodent liver tumors
An important application of transcriptomics tools has been developing an understanding of the mode of action of suspected “non-genotoxic carcinogens”. Physiological and biochemical differences between humans and rodents mean that the same compound might affect different species in different ways. Because most decisions about human safety are made based on experiments using rodents, understanding these differences is essential.
Compounds that cause tumors in rodents by activation of a particular class of signaling pathways (hepatic nuclear receptors such as peroxisome proliferator activated receptor alpha (PPARα) and constitutive androstane receptor (CAR)) are of questionable relevance to human health. Many commercially relevant compounds are known to operate by this mode of action, but the list of these compounds is not exhaustive. Many compounds commonly used in consumer products, building materials, and other media associated with near-field human exposures have been shown to interact with hepatic nuclear receptors. For example, phthalates such as di(2-ethylhexyl)phthalate are readily found in food packaging1 and have unambiguously been associated with activation of PPARα2. The series of events (i.e., adverse outcome pathways) from binding of a chemical to PPARα to rodent hepatic tumors has also been well established4-6. However, while this mode of action is well described in rats, the adverse endpoint is irrelevant to humans. Acute and chronic exposures to PPARα agonists are not associated with tumors in humans or non-human primates5. Based on observed differences in species susceptibility, the International Agency for Research on Cancer has categorized DEHP as a Class 3 carcinogen (no evidence of cancer causing potential in humans)7.
An application of the MoAviz tools for interpreting transcriptomic data
ScitoVation has developed an interactive visual platform called MoAviz to make better use of transcriptomic data for understanding compound mode of action. To illustrate the utility of transcriptomic studies for understanding compound mode of action, we considered an existing transcriptomic dataset that involved treatment of several mouse strains to di(2-ethylhexyl)phthalate (DEHP)3. In this study, wild-type (129S1/SvImJ) and PPARα-null mice were treated daily with DEHP via gavage for four days. Gene expression in the liver was assessed for both genotypes relative to control. 475 genes were significantly altered in wild-type animals, versus 190 genes in PPARα-null animals.
Based on this data, we prepared a map of the functional changes in the liver in response to DEHP in the presence and absence of PPARα (Figure 1). We determined which Gene Ontology (GO) terms were overrepresented among the genes up- and down-regulated in response to DEHP treatment. Connecting GO terms based on their hierarchical definitions provides a quick snapshot of the gene regulatory program imparted by a perturbation. Here, we see that fatty-acid, cholesterol, and coenzyme metabolism are all altered by DEHP in wild-type animals (Figure 1A). These are classical PPARα-induced pathways, similar to what we observed in-house with rats treated with the selective PPARα agonist GW7647. These responses are nearly completely absent in the PPARα-null mice. The similarity of the transcriptional profile to that of a pure PPARα agonist, combined with the lack of response in the knockout, is strong evidence for a PPARα-mediated mode-of-action
References
- K. M. Rodgers, et al., in Toxicants in Food Packaging and Household Plastics: Exposure and Health Risks to Consumers, ed. S. M. Snedeker, Springer London, London, Editon edn., 2014, pp. 31-59.
- C. H. Hurst, et al., Toxicological sciences : an official journal of the Society of Toxicology, 2003, 74, 297-308.
- H. Ren, et al., Toxicological sciences : an official journal of the Society of Toxicology, 2010, 113, 45-59.
- R. C. Cattley, Toxicol Pathol, 2004, 32 Suppl 2, 6-11.
- R. C. Cattley, et al., Regul Toxicol Pharmacol, 1998, 27, 47-60.
- J. E. Klaunig, et al., Crit Rev Toxicol, 2003, 33, 655-780.
- IARC Monographs, Di(2-ethylhexyl) phthalate (DEHP), monographs.iarc.fr/ENG/Publications/techrep42/TR42-18.pdf (May 4, 2017).
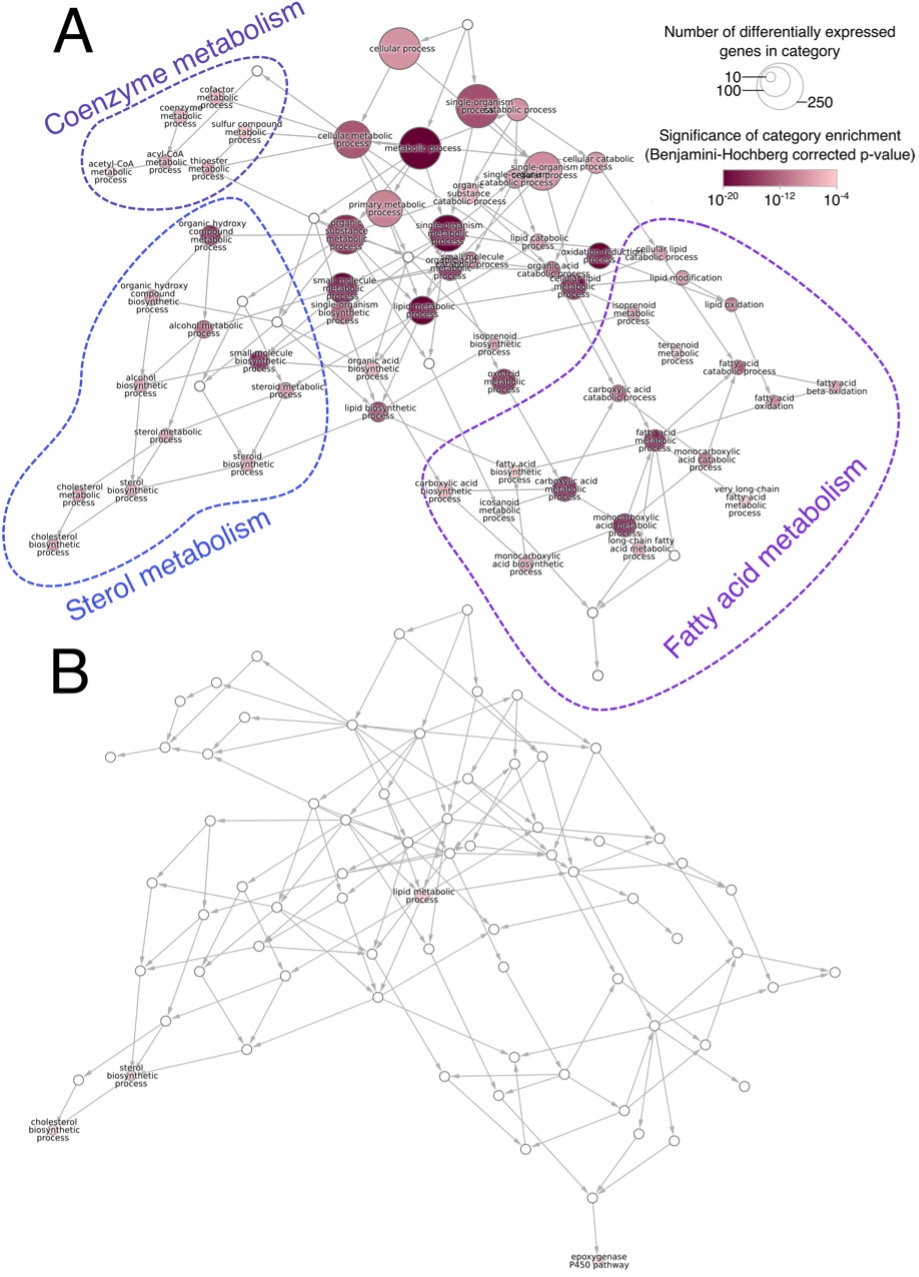
Figure 1. The network of Gene Ontology (GO) biological processes enriched genes upregulated by DEHP treatment in (A) wild-type and (B) PPARα-null mice. The number of genes annotated with each term is reflected by node size and the significance of the enrichment by node color (hypergeometric test, GO terms with Benjamini-adjusted p<10‑4 shown). Non-significant nodes were included (white) as needed to complete the minimal spanning tree. Consistent with a PPARa-activation mode-of-action, DEHP has a profound effect on lipid and carboxylic acid metabolism processes. This response is almost completely absent in the PPARα-null animals.
.
